Machine Learning Model Selection and Validation
Overview
Teaching: 30 min
Exercises: 30 minQuestions
How can we be sure that our model is performing as well as we think?
What are some techniques to overcome common ML issues?
Objectives
Apply a Student’s t-test to validation accuracies.
Overcome challenges posed by imbalanced data.
In the last session we trained a classification model on some micrscope imagery conatin protein crystals. Towards the end of the session we applied a range of different performance metrics that illuminated different aspects of the models performance. Ultimately we observed that the model was heavily biased by the unbalanced distribution of classes in our training set. How do we know this wasn’t just by chance? Can we get some statistical guarantee about our findings?
Single Model Validation
A nice way to test statistical significance is through the use of cross validation. For each fold of validation we can measure an accuracy (or some other metric of interest), thus with a sample of accuracies we can now estimate a distribution over accuracies. With a distribution of accuracies we can now apply a Student’s t-Test to test for a statistically significant difference from some benchmark value. We can do it all with a few lines of Python code. In case your environment has reset, or you are coming in to this session raw, let’s just redownload or data and define our classifier.
from utils.datasets import c3
(X_train, y_train), (X_test, y_test) = c3.load_data()
Downloading datafile to /root/data/crystals.npy ...
... done
Downloading datafile to /root/data/clear.npy ...
... done
Sub-sampling dataset...
... shuffling and splitting
... done
Authentication
The first time you execute one of the dataset utility functions you will be asked to authenticate with you google credentials, don’t be afriad.
Go to the following link in your browser: https://accounts.google.com/o/oauth2/auth?redirect_uri=..... Enter verification code:
from sklearn import svm
classifier = svm.SVC(gamma=0.001, verbose=True)
SVC(C=1.0, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape='ovr', degree=3, gamma=0.001, kernel='rbf',
max_iter=-1, probability=False, random_state=None, shrinking=True,
tol=0.001, verbose=True)
Cross Validation
Cross validation can be implemented using scikit-learn in a single line
from sklearn.model_selection import cross_validate
cv_results = cross_validate(classifier, X_train, y=y_train, cv=10, verbose=3, n_jobs=-1)
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 2 concurrent workers.
[Parallel(n_jobs=-1)]: Done 10 out of 10 | elapsed: 5.3min finished
The above code will perform a 10-fold cross-validation on our traing data set. You can specify specific metrics that you would like to collect across the folds, but there are some default values chosen for you
print(cv_results.keys())
dict_keys(['fit_time', 'score_time', 'test_score', 'train_score'])
For our t-Test we are interested in the test_score metric, which in this case is simply the accuracy. We can now calculate the one-sided t-statistic for our accuracy and some benchmark value, lets say an accuracy of 80%.
from scipy import stats
accuracies = cv_results["test_score"]
k = len(accuracies)
benchmark = 0.8
m = np.mean(accuracies)
S = np.std(accuracies)
SE = S / np.sqrt(k)
t = (m - benchmark) / SE
p = stats.t.cdf(t,df=2*k - 2)
print("The accuracy of you model is %.2f\u00B1%.2f" %(m, 2.58*SE))
print("The p-value for this test is %.3f" % p)
The accuracy of you model is 0.80±0.00
The p-value for this test is 0.103
In this case it looks like we cannot say with confidence that our model accuracy is greater than or equal to 80%.
Resampling
So, we have identified that our dataset is imabalanced and is causing our model to underperform. How can we fix this problem? A simple way is to resample our dataset, that is oversample the minority class or undersample the majority class. There is a nice project dedicated to providing easy to use solution to such problems, imbalanced-learn. It can both over and under sample datasets for us with relative ease. Let’s start with over sampling
from sklearn.utils import shuffle
from imblearn.over_sampling import RandomOverSampler
ros = RandomOverSampler()
X_oversampled, y_oversampled = ros.fit_resample(X_train, y_train)
X_oversampled, y_oversampled = shuffle(X_oversampled, y_oversampled)
print("Fraction of samples assigned to the crystal class %.2f%%" % (np.sum(y_oversampled) / y_oversampled.shape[0] * 100.))
Fraction of samples assigned to the crystal class 50.00%
Too easy. Let’s run our cross-validation analysis again and see if this approach improves the model at all.
classifier = svm.SVC(gamma=0.001)
cv_results = cross_validate(classifier, X_oversampled, y=y_oversampled, cv=10, verbose=3, n_jobs=-1)
accuracies = cv_results["test_score"]
k = len(accuracies)
benchmark = 0.8
m = np.mean(accuracies)
S = np.std(accuracies)
SE = S / np.sqrt(k)
t = np.abs((m - benchmark) / SE)
p = 1. - stats.t.cdf(t,df=2*k - 2)
print("The accuracy of you model is %.2f\u00B1%.2f" %(m, 2.58*SE))
print("The p-value for this test is %.3f" % p)
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 2 concurrent workers.
/usr/local/lib/python3.6/dist-packages/sklearn/externals/joblib/externals/loky/process_executor.py:706: UserWarning: A worker stopped while some jobs were given to the executor. This can be caused by a too short worker timeout or by a memory leak.
"timeout or by a memory leak.", UserWarning
[Parallel(n_jobs=-1)]: Done 10 out of 10 | elapsed: 10.2min finished
The accuracy of you model is 0.99±0.00
The p-value for this test is 0.000
Wow! Well it has certainly improved our accuracy, with a statistcally significant difference from our benchmark value, but something is not right here. We have a confidence interval of +- 0.? That seems a little suspicious. Let’s run some of the performance metrics we covered in the previous section.
predicted = classifier.predict(X_test)
print("Classification report for classifier %s:\n%s\n"
% (classifier, metrics.classification_report(y_test, predicted)))
Classification report for classifier SVC(C=1.0, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape='ovr', degree=3, gamma=0.001, kernel='rbf',
max_iter=-1, probability=False, random_state=None, shrinking=True,
tol=0.001, verbose=False):
precision recall f1-score support
0.0 0.00 0.00 0.00 123
1.0 0.80 1.00 0.89 477
micro avg 0.80 0.80 0.80 600
macro avg 0.40 0.50 0.44 600
weighted avg 0.63 0.80 0.70 600
Hmmmm. We seem to have gone back to square one. Why are we not performing so well on our test set? We were getting 99% accuracy on our cross-validation run above. This is a classic indicator of over-fitting. What we are seeing here is an extreme difference between our training accuracy and our test accuracy, a typical red flag for over-fitting.

Over-fitting is essentially the point were your model starts remembering individual data points instead of learning generalisable patterns. I think the presence of the zero variance in the cross-validation accuracy and the large divergence of the test and train accuracies are dead give aways that we have overfit. This is most likely due to the fact that we have oversampled the minority class. Given the 4:1 ration of crystal to clear images, each clear image would be, on average, sampled four times. So, perhaps the model has learned individual clear samples (having been shown them many more times than the crystal images) and has learned to detect some features of the crystal class. Thus when it is shown clear images it hasn’t seen before it does not recognise them as clear.
Let’s try the alternative, let;s undersample.
from imblearn.under_sampling import ClusterCentroids
cc = ClusterCentroids(random_state=0)
X_resampled, y_resampled = cc.fit_resample(X_train, y_train)
X_resampled, y_resampled = shuffle(X_resampled, y_resampled)
classifier = svm.SVC(gamma=0.001)
cv_results = cross_validate(classifier, X_oversampled, y=y_oversampled, cv=10, verbose=3, n_jobs=-1)
accuracies = cv_results["test_score"]
k = len(accuracies)
benchmark = 0.8
m = np.mean(accuracies)
S = np.std(accuracies)
SE = S / np.sqrt(k)
t = np.abs((m - benchmark) / SE)
p = 1. - stats.t.cdf(t,df=2*k - 2)
print("The accuracy of you model is %.2f\u00B1%.2f" %(m, 2.58*SE))
print("The p-value for this test is %.3f" % p)
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 2 concurrent workers.
[Parallel(n_jobs=-1)]: Done 10 out of 10 | elapsed: 51.5s finished
The accuracy of you model is 0.51±0.01
The p-value for this test is 0.000
So now we are back to random guessing? It doesn’t look like our model is really having a good time here.
predicted = classifier.predict(X_test)
print("Classification report for classifier %s:\n%s\n"
% (classifier, metrics.classification_report(y_test, predicted)))
Classification report for classifier SVC(C=1.0, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape='ovr', degree=3, gamma=0.001, kernel='rbf',
max_iter=-1, probability=False, random_state=None, shrinking=True,
tol=0.001, verbose=False):
precision recall f1-score support
0.0 1.00 0.01 0.02 123
1.0 0.80 1.00 0.89 477
micro avg 0.80 0.80 0.80 600
macro avg 0.90 0.50 0.45 600
weighted avg 0.84 0.80 0.71 600
One Last Model
Okay. One last try. Let’s see what a decision tree can do.
from sklearn import tree
classifier = tree.DecisionTreeClassifier()
classifier.fit(X_train, y_train)
expected = y_test
predicted = classifier.predict(X_test)
print("Classification report for classifier %s:\n%s\n"
% (classifier, metrics.classification_report(expected, predicted)))
plot_confusion_matrix(y_test, predicted, classes=np.array(["Clear", "Crystal"]))
cv_results = cross_validate(classifier, X_train, y=y_train, cv=10, verbose=3, n_jobs=-1)
accuracies = cv_results["test_score"]
k = len(accuracies)
benchmark = 0.8
m = np.mean(accuracies)
S = np.std(accuracies)
SE = S / np.sqrt(k)
t = np.abs((m - benchmark) / SE)
p = 1. - stats.t.cdf(t,df=2*k - 2)
print("The accuracy of you model is %.2f\u00B1%.2f" %(m, 2.58*SE))
print("The p-value for this test is %.3f" % p)
Classification report for classifier DecisionTreeClassifier(class_weight=None, criterion='gini', max_depth=None,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, presort=False, random_state=None,
splitter='best'):
precision recall f1-score support
0.0 0.67 0.68 0.68 123
1.0 0.92 0.91 0.92 477
micro avg 0.87 0.87 0.87 600
macro avg 0.79 0.80 0.80 600
weighted avg 0.87 0.87 0.87 600
Confusion matrix, without normalization
[[ 84 39]
[ 41 436]]
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 2 concurrent workers.
[Parallel(n_jobs=-1)]: Done 10 out of 10 | elapsed: 38.6s finished
The accuracy of you model is 0.85±0.02
The p-value for this test is 0.000
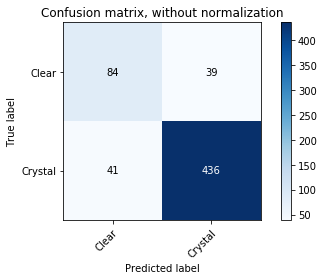
We seem to have improved in every aspect here. We have a respectable accuracy that is statistically distinct from the benchmark value. We seem to have decreased the effects of overfitting while still maintaining a high accuracy. Yay decision trees!
Key Points
Traditional statistical methods for validation are still extremely useful.
Most of the time your data is much more important that the model itself.